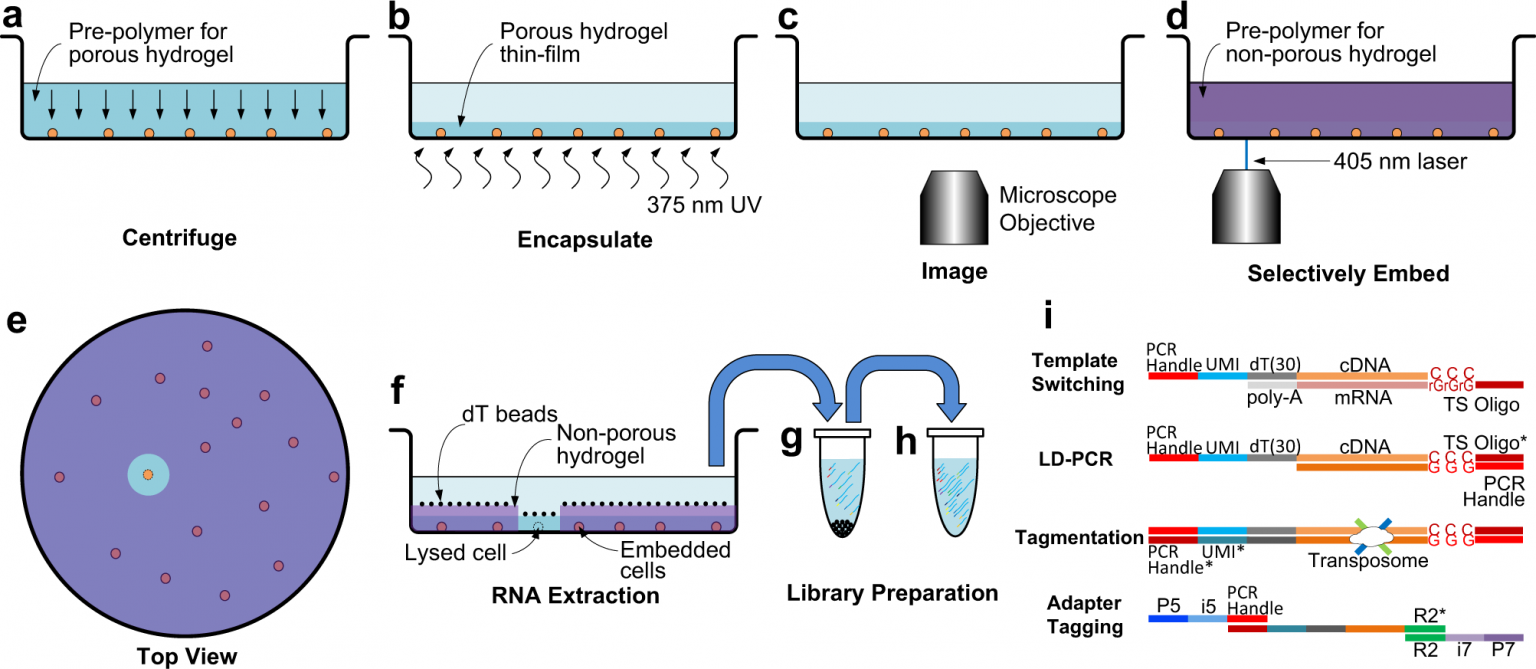
The Multi Scale Design Laboratory (MDL) lead by Professor Hongshen Ma has recently published a new paper, “See-N-Seq: RNA sequencing of target single cells identified by microscopy via micropatterning of hydrogel porosity”. MDL researchers have developed a new method, “See-N-Seq,” that uses a micropatterned hydrogel to capture target cells for single cell RNA sequencing. This approach enables rapid screening of cells without having to physically transport individual cells to prepare them for sequencing.
Single cell RNA sequencing can provide unique insights into the genes that produce specific cellular phenotypes and behaviours. However, these observable traits are often elusive or can only be identified using imaging, which makes selecting cells at random for sequencing an ineffective technique. See-N-Seq provides an efficient method to isolate specific single cells from a larger sample. The sample material is placed in standard microwell plates, where an initial layer of porous hydrogel traps the sample cells so they can be analyzed at the same time through microscopy. Once the target cell is identified, a second layer of hydrogel – this time non-porous and micropatterned with a laser – traps the other cells while allowing the target cell to be accessed and allowing in-place RNA extraction. This method allows genetic data to be associated with specific cellular phenotypes and behaviours.
“See-N-Seq: RNA sequencing of target single cells identified by microscopy via micropatterning of hydrogel porosity” has been published in Communications Biology, part of the Nature portfolio of journals. It is available at DOI: https://doi.org/10.1038/s42003-022-03703-3.